RASREC Rosetta
This chapter describes the RASREC protocol as described in Refs. [1][2]. RASREC is an iterative conformational sampling protocol that seeks to pool knowledge gained about the conformational space in previous trajectories to efficiently guide further space exploration. The protocol is inherently parallel and requires inter process communication which is realized with MPI. RASREC runs relatively independent structure calculations such that a low-latency, low-bandwith connectivity is sufficient. Since RASREC builds on the basic CS-Rosetta protocol options and input-files for the basic CS-Rosetta protocols are also relevant here. The protocol can be characterized in the following way: 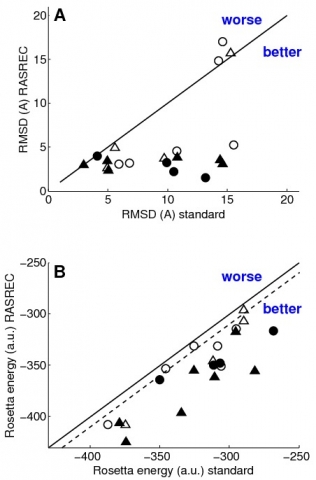
- fragment library is picked using chemical shift data
- individual structure calculations use the abrelax sampling protocol
- 200-1000 of independent structure calculations run in parallel
- a pool of the best output decoys is continuously updated from the results of the independent structure calculations
- features in the pooled structures are used to focus further sampling
Generally we found that RASREC improves convergence of CS-Rosetta and is superior in exploiting experimental restraint information, as shown in the Figure to the right.
Preparation of Input Files
RASREC uses the same input files as the basic CS-Rosetta protocol.
Running RASREC CS-Rosetta
To prepare a RASREC simulation perform the same setup steps as described before but use setup_target -method rasrec for your method-choice. RASREC runs parallel structure calculations in batches using its own work-load manager. Sub-directories 'batch_000001', 'batch_000002', 'batch_000003' are automatically generated when a new batch is started and are populated with all necessary input files, e.g., batch-dependent flags, broker setup and structural input information. The calculated structures of a batch are generally stored in batch_nnnnn/decoys.out. Intermediate structures after completion of abinitio-stages (stage1, stage2, stage3 and stage4) are stored in decoys_stage1.out,..., decoys_stage4.out, accordingly. These intermediate decoys are required for the IVth resampling stage of the RASREC protocol, which performs proto-fold resampling[2].
A work-load manager watches the completion of structure calculations in current batches and presents final structures to the global structural pool. The best scoring structures are retained in the pool and all others are discarded. The size of the pool is controlled by flag
RASREC uses 6 resampling stages I, II, ... VI, which differ in the way how the structural information in the pool is used for resampling and whether well-refined full-atom structures are produced or just low-resolution (centroid) models. For details see Ref.
The pool directory (called centroid_pool during stages I-IV and fullatom_pool during stages V-VI) contains the files decoys.out, decoy.out.backup and STATUS. The set of structures in decoys.out always reflects the current lowest 500 (-iterative:pool_size) decoys. The ScoreFunction to select these 500 is different from the ScoreFunction used for sampling. This allows to up-scale restraints for selection without distorting the structures during sampling. Moreover, it allows to include scores (like the chemical shift score) that are not computed during sampling. The ScoreFunction for the low-energy decoy pool can be patched using flags -iterative:cen_score_patch and -iterative:fa_score_patch for centroid and fullatom pool respectively. The silent file score-column "_archive_select_score_" contains the combined weight according to the pool ScoreFunction and is the number used for acceptance and rejection into the pool. To avoid bottlenecks this combined score is actually pre-computed within the batches. In some cases the presence of all-atoms in the pooled structures is required. This is for instance during automatic NOESY assignment and chemical shift rescoring the case. In these cases RASREC also stores full-atom models in the centroid pool. However, these structures have only seen very rudimentary refinement such that their full-atom energy scores are not useful for decoy discrimination. Instead the pre-all atom centroid score is stored in silent file score-column 'prefa_centroid_score'.
Restarting RASREC calculations
RASREC calculations can be restarted without the need of further interaction when stopped prematurely. The program will find all necessary status information in the files centroid/fullatom_pool/STATUS and in the BATCH_INFO files for the individual batches. When the program is stopped all the work in unfinished trajectories will be lost in the same manner as for the CS-Rosetta protocol. Hence also for RASREC high frequencies of preemption in comparison to the average trajectory length would lead to significant loss of effectiveness.
Rarely the crash of the program leaves files corrupted. In these cases it helps to delete the newest batch_nnnn directories (which are often still empty). Sometimes the centroid/fullatom_pool/decoys.out file is corrupted. In this case one can manually copy the decoys.out.backup file back to decoys.out.
Post-Processing of RASREC structure calculations
A RASREC simulation is finished if directory fullatom_pool_stage8 exists. The final decoys will be in silent file fullatom_pool/decoys.out. Usually it is enough to extract the lowest energy structures from this low-energy pool. If the analysis requires an overview over all produced structures one has to note that decoy tags although unique within each batch_000nnn/decoys.out file are duplicated across different batches. Thus, it is best to use the application collect_decoys XXX write it as XXX to combine the decoys from the batch files.
For more information see the respective Tutorials and the Application Reference.
References
- [raman2010a] .
2010. NMR structure determination for larger proteins using backbone-only data.. Science. 327(5968):1014-1018. - [Lange2012a] .
2012. Resolution-adapted recombination of structural features significantly improves sampling in restraint-guided structure calculation.. Proteins. 80(3):884-895.