Computational Requirements
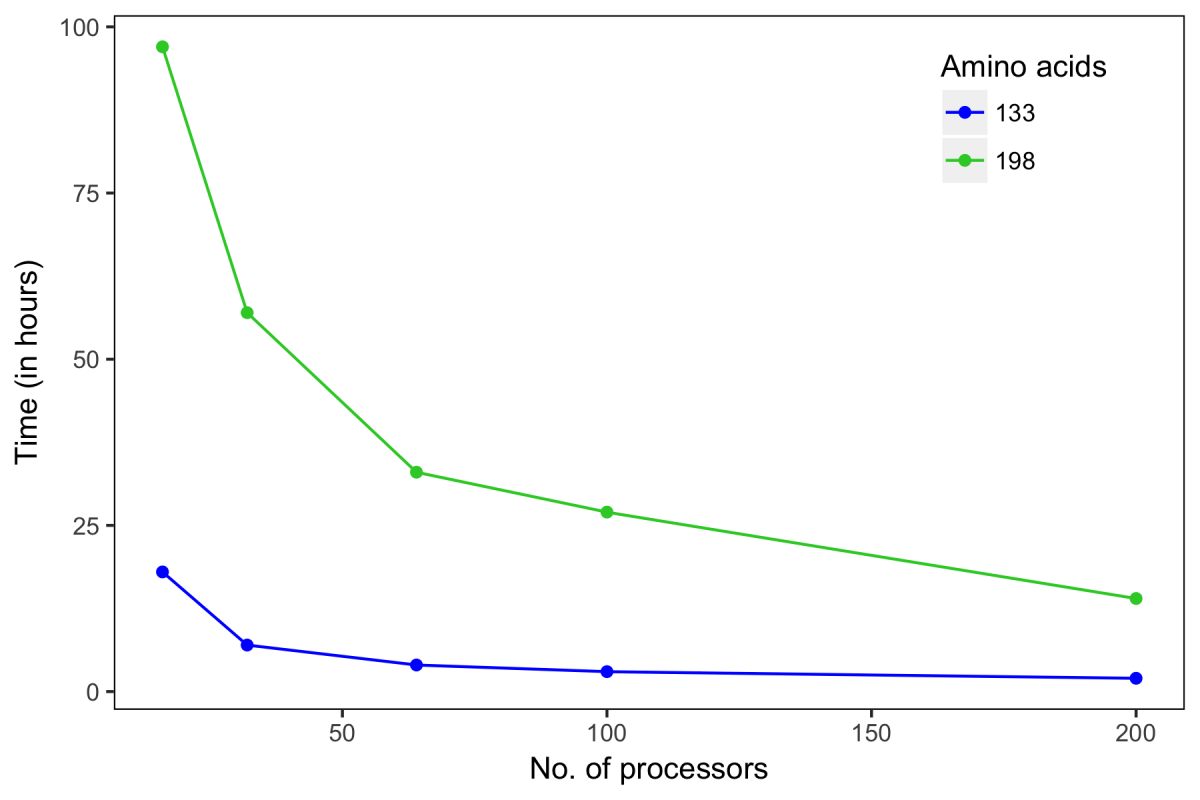
Typical RASREC and AutoNOE structure calculations require a computer cluster consisting of 200-2000 processors (or cores). However, they can be run on any Unix-based cluster with 16 or more parallel processors. The number of processors used for structure calculations directly influence the performance of these protocols. For instance, to calculate the structure of a protein with 100 amino acids, RASREC/AutoNOE would typically require ~6 to 8 hours on a cluster of 100 processors. The time required for the same setting with smaller number of clusters (say 16 or 32), would be a couple (~3 to 4) of days (see the plot below). Whereas, standard CS-Rosetta protocol can still be used on any commodity computer.
Below we show the performance measure of CS-Rosetta's AutoNOE protocol represented as a function of time and number of processors. To evaluate this measure, we performed the structure calculations of two targets, Regulator of Ty1 Transposition (RTT, 133 amino acids) and α-lytic protease (aLP, 198 amino acids). Here, we used as input, chemical shifts assigned by 4D-CHAINS[1] and two 4D NOESY (HCCH and HCNH) unassigned peak lists. The points on the plot represent independent structure calculations of respective targets (indicated by colors) using specific set of processors (x-axis; processors used: 16, 32, 64, 100, 200). All these calculations were setup to produce 100 structures in every batch. Depending on the number of processors used, the calculations took ~14-97 hours for aLP and ~2-7 hours for RTT.